Author: Savannah Sadaiappen
The information used in this post can be found in the article, “Development of a low-cost and portable smart fluorometer for detecting breast cancer cells” by Mohammad Wajih Alam, Khan A. Wahid, Raghuveera Kumar Goel, and Kiven Erique Lukong, published in January 2019.
I’m sure we have all heard of the fascinating green fluorescent protein, commonly known as GFP, at least once and thought the idea of it was the coolest thing ever right? It’s basically like making the cells you’re researching glow and who doesn’t love that? It’s essentially a party…under a microscope #lit. However, the use of GFP and detecting it via fluorescence microscopy can cost scientists a lot of dolla dolla bills y’all (it’s expensive).

https://giphy.com/gifs/migos-james-corden-carpool-karaoke-1hMhl5fpuLldDqj0Ey
Not all scientists have as much money as Takeoff (member of Migos) and unfortunately aren’t able to utilize fluorescence imaging instruments. Scientists at the University of Saskatchewan researched the development of a low-cost and portable smart fluorometer to combat this issue.
Fluorescence microscopy is based on the principle of fluorescence imaging which serves as a powerful tool in the fields of biology and chemistry to monitor cell dynamics and molecules. Commercial fluorescent microscopes have numerous advantages: live cell imaging, wide field-of-view, and sensitive sophisticated cameras for high resolution images to name a few. By fluorescently labelling proteins of interest scientists can observe/investigate any cellular process and use this to aid in the extraction of meaningful information.

https://www.bioimager.com/microscope-fun/
Fluorescence microscopy has major applications in cancer biology where cancer cells are chemically labeled for detection. As breast cancer is the most common form of cancer affecting women with 1 in 8 women expected to be diagnosed with the diseases during her lifetime the development of a low cost and portable detection instrument offers numerous advantages to breast cancer research. The research described in the article was used to design a low-cost and portable fluorometer that can be used to detect fluorescence signals emitted from a model breast cancer cell line which have been engineered to express the green fluorescent protein (GFP). The device used utilizes a flashlight that works in the visible range which serves as an excitation source and a photodiode as a detector. It also uses an emission filter which mainly allows the fluorescence signal to reach the detector while eliminating the use of an excitation filter and dichroic mirror. The emission filter was selected so that it matches the emission wavelength if GFP which ensured that the GFP wavelength is the main wavelength allowed to pass through. The elimination of these two complicated devices is what makes the newly developed instrument compact, low-cost, portable, and lightweight. The set up of the device can be seen below in Figure 1.
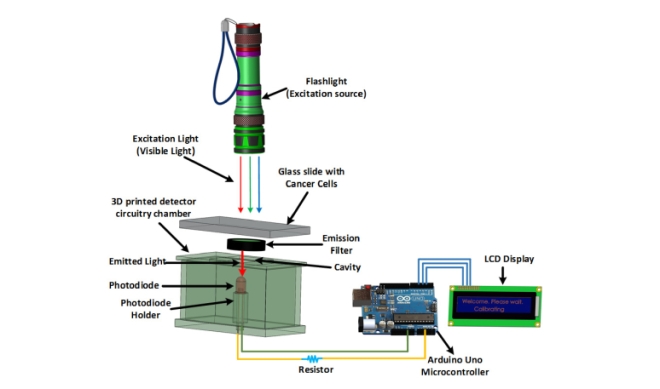
Fig 1. A graphical illustration and working principle of the developed device.
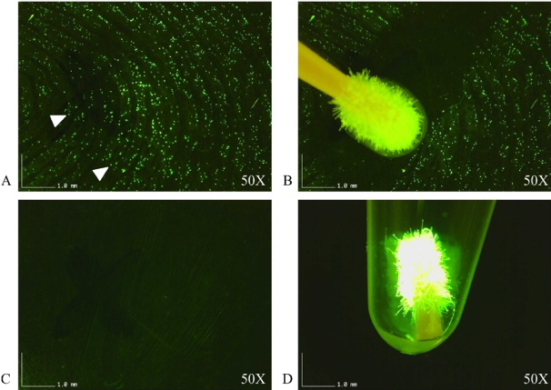
The device developed consists of a flashlight which works in visible range, a photodiode to detect the fluorescent signal, a microcontroller to read and transmit the data, and an LCD screen to display messages as a major component. Immortalized human breast cancer cells were used to test the fluorometer prototype. The gene encoding the Green Fluorescent Protein (GFP) was stably introduced into these cells via retroviral transduction. GFP expression of these live cells was confirmed via fluorescence microscopy which served as a control. These same samples were used to measure the intensity of the fluorescence signal using the developed fluorometer. A schematic of how the device works can be seen in figure 2.
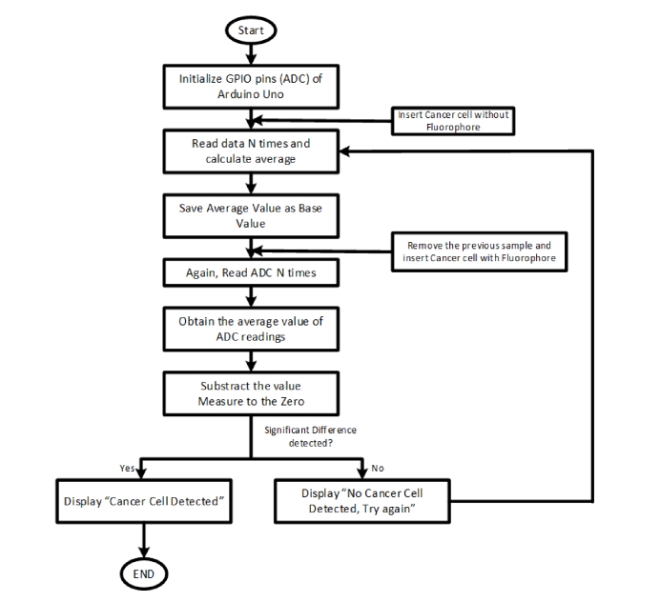
Fig 2. Operating procedure of the system
Detection of GFP in the cancer cells was successful and confirmed the development of the prototype fluorometer was successful after scientists analyzed statistics and accuracy of the device. The development of this low cost fluorometer which can measure a signal emitted by breast cancer cells expressing GFP promotes the access of fluorometers to medical and research laboratories where resources are limited. This fluorometer has broad applications in the fluorescence-based detection of multiple cancer types and can greatly impact research done by the clinical and biomedical community.

https://giphy.com/gifs/nickcannon-nick-cannon-MSCzxrLEF25feISTIz
Article can be found at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6377908/ (figures also sourced from this article)